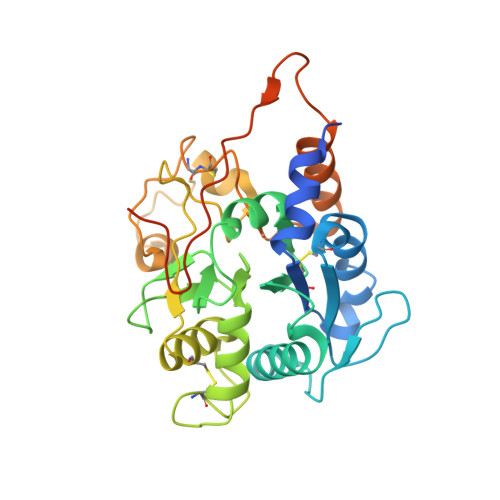
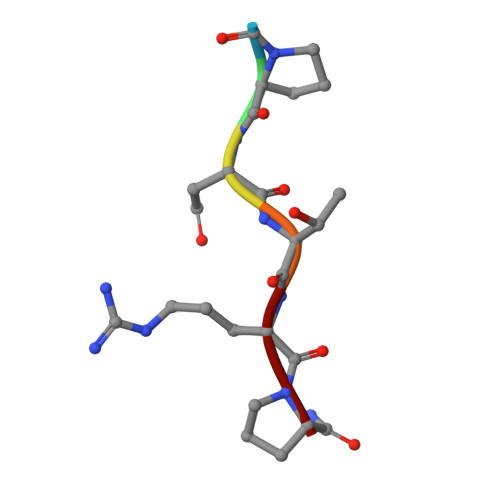
Structural basis for the synthesis of the core 1 structure by C1GalT1.
Gonzalez-Ramirez, A.M., Grosso, A.S., Yang, Z., Companon, I., Coelho, H., Narimatsu, Y., Clausen, H., Marcelo, F., Corzana, F., Hurtado-Guerrero, R.(2022) Nat Commun 13: 2398-2398
- PubMed: 35504880
- DOI: https://doi.org/10.1038/s41467-022-29833-0
- Primary Citation of Related Structures:
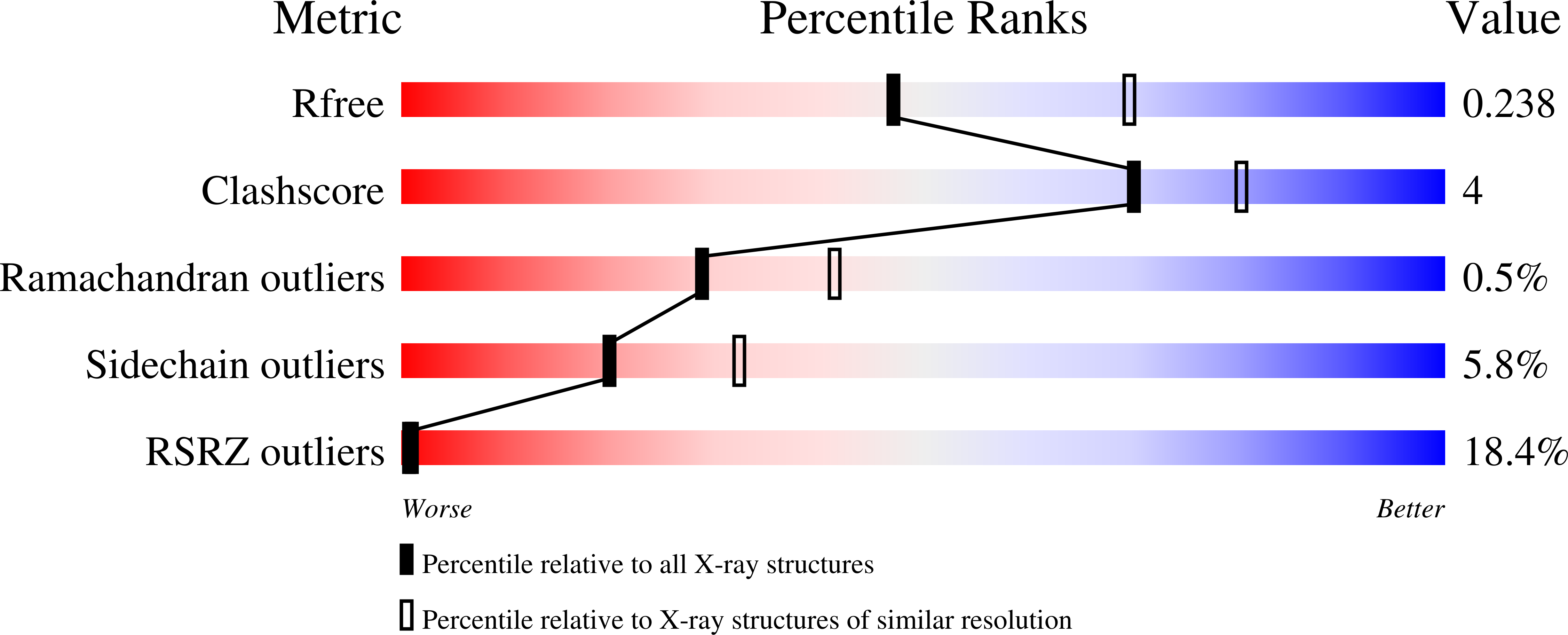
7Q4I - PubMed Abstract:
C1GalT1 is an essential inverting glycosyltransferase responsible for synthesizing the core 1 structure, a common precursor for mucin-type O-glycans found in many glycoproteins. To date, the structure of C1GalT1 and the details of substrate recognition and catalysis remain unknown. Through biophysical and cellular studies, including X-ray crystallography of C1GalT1 complexed to a glycopeptide, we report that C1GalT1 is an obligate GT-A fold dimer that follows a S N 2 mechanism. The binding of the glycopeptides to the enzyme is mainly driven by the GalNAc moiety while the peptide sequence provides optimal kinetic and binding parameters. Interestingly, to achieve glycosylation, C1GalT1 recognizes a high-energy conformation of the α-GalNAc-Thr linkage, negligibly populated in solution. By imposing this 3D-arrangement on that fragment, characteristic of α-GalNAc-Ser peptides, C1GalT1 ensures broad glycosylation of both acceptor substrates. These findings illustrate a structural and mechanistic blueprint to explain glycosylation of multiple acceptor substrates, extending the repertoire of mechanisms adopted by glycosyltransferases.
Organizational Affiliation:
Institute of Biocompuation and Physics of Complex Systems, University of Zaragoza, Mariano Esquillor s/n, Campus Rio Ebro, Edificio I+D, 50018, Zaragoza, Spain.